-
Notifications
You must be signed in to change notification settings - Fork 8
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Add Report from the Helmholtz Metadata Collaboration Conference (HMC)…
… 2024 for group2
- Loading branch information
1 parent
b79e923
commit 5d97939
Showing
1 changed file
with
76 additions
and
0 deletions.
There are no files selected for viewing
76 changes: 76 additions & 0 deletions
76
posts/feed_bot/galaxy_europe/2024-11-08-galaxy-imaging-fair-pipelines-group2.md
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,76 @@ | ||
| --- | ||
| media: | ||
| - bluesky-galaxyproject | ||
| mentions: | ||
| bluesky-galaxyproject: | ||
| - galaxyproject.bsky.social | ||
| hashtags: | ||
| bluesky-galaxyproject: | ||
| - UseGalaxy | ||
| - GalaxyProject | ||
| - UniFreiburg | ||
| - EOSC | ||
| - EuroScienceGateway | ||
| --- | ||
| 📝 New blog post Released! | ||
| https://galaxyproject.org/news/2024-11-08-galaxy-imaging-fair-pipelines/ | ||
|
|
||
| The Helmholtz Metadata Conference (HMC) 2024 | ||
| -------------------------------------------- | ||
|
|
||
| The Helmholtz Metadata Collaboration (HMC) focuses on enhancing research | ||
| data quality through metadata, implementing this approach throughout the | ||
| entire organisation. Its primary goal is to make the extensive research | ||
| data produced by Helmholtz Centres findable, accessible, interoperable, | ||
| and reusable (FAIR) for the scientific community. This year’s Helmholtz | ||
| Metadata Conference (HMC) was held virtually in Gather Town from November | ||
| 4th to 6th. The event targeted scientists, data professionals, software developers, | ||
| and anyone engaged with metadata. Sessions covered a range of topics, | ||
| including metadata management, ontologies, metadata schemas and formats, | ||
| semantics, standardisation, and workflows. | ||
|
|
||
|  | ||
|
|
||
| Building FAIR Bioimage Pipelines with Galaxy | ||
| -------------------------------------------- | ||
|
|
||
| In the “workflow” session, I presented the application of Galaxy to develop | ||
| FAIR analysis pipelines for metadata annotation and image management in | ||
| high\-content screening (HCS) bioimaging. In fact, Galaxy workflows enhance | ||
| data FAIRness by allowing annotation, collaboration, and public sharing. | ||
| Additionally, Galaxy tools are transparent, meaning they can be easily | ||
| inspected within the Galaxy Tool Shed, providing a clear view of their | ||
| structure. | ||
|
|
||
|  | ||
|
|
||
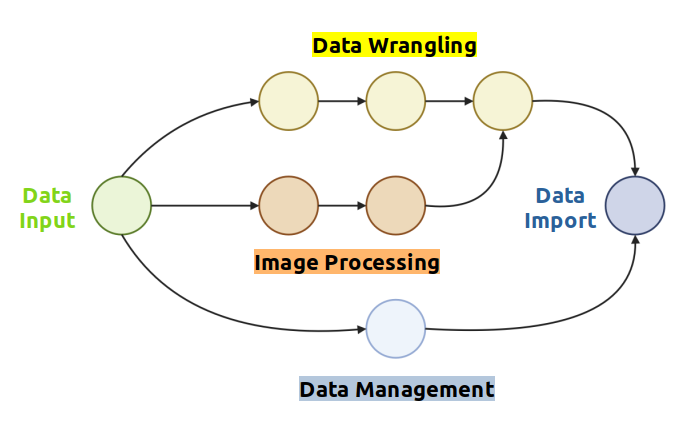
| Integrating OMERO and Galaxy | ||
| ---------------------------- | ||
|
|
||
| During the conference, I discussed integrating OMERO’s upload and image | ||
| annotation features with Galaxy’s image analysis workflows. | ||
| This integration enables images to be automatically enriched with metadata | ||
| (e.g., key\-value pairs, tags, raw data, regions of interest) and uploaded | ||
| to a user\-defined OMERO server using a new OMERO tool suite developed in | ||
| Galaxy. Currently, the suite offers tools for image and ROI uploads, | ||
| metadata enrichment, and data filtering or retrieval, making it simple | ||
| to transfer data and metadata to an OMERO instance. | ||
|
|
||
|  | ||
|
|
||
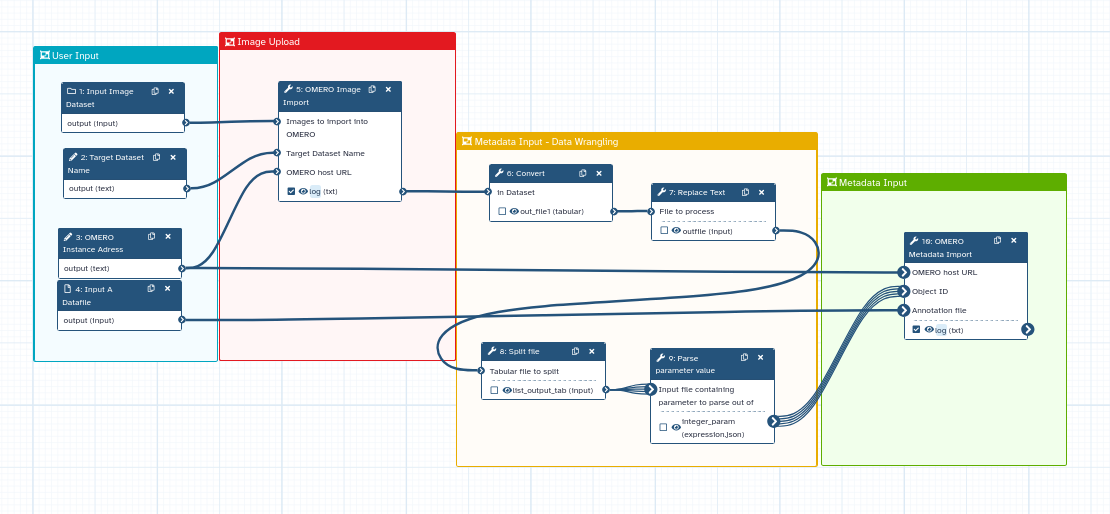
| Combining Image Processing and Data Management in One Workflow | ||
| -------------------------------------------------------------- | ||
|
|
||
| Image processing can be added to OMERO workflows. Users can retrieve | ||
| images from a local server and perform image analyses, such as annotation, | ||
| segmentation, or background subtraction. Galaxy is a powerful tool for | ||
| image analysis, integrating popular software like CellProfiler and connecting | ||
| with cloud services like OMERO and the IDR. This integration allows for | ||
| efficient access to and management of image data, which is invaluable in | ||
| bioimaging. Automated pipelines streamline the handling of complex metadata, | ||
| ensuring data integrity and fostering interdisciplinary collaboration. | ||
| This approach not only boosts the efficiency of HCS bioimaging but | ||
| also aligns with the scientific community’s FAIR principles, driving | ||
| scientific discovery and innovation forward. | ||
|
|
||
| The presentation was shared on [Zenodo](https://zenodo.org/records/14044640) |