-
Notifications
You must be signed in to change notification settings - Fork 587
How mrjob works
This is a brief walkthrough of the mrjob codebase. It's aimed primarily at mrjob maintainers and contributors, but should also be useful for anyone trying to debug an issue with mrjob.
mrjob lets you describe a job in Python, and then helps you run it on Spark or Hadoop Streaming, as painlessly as possible. Some pain points that mrjob relieves are:
- dealing with Elastic MapReduce and other cloud services; for example creating temp space on S3, setting up IAM instance profiles and service roles, and dealing gracefully with throttling by the EMR API
- making sure job can access libraries and other files it depends on, and that they are correctly initialized (e.g. running
make) - if your job fails, helping you dig the error out of logs
In mrjob, jobs are always a subclass of the MRJob class (found in mrjob/job.py). Jobs do three things:
- read arguments from the command line
- set up a runner and tell it to run the job
- run inside Spark or Hadoop Streaming
A mrjob script is just a Python file containing a definition of a MRJob subclass, and a small snippet of code at the end ensuring that if you run the script, the run() method on that subclass gets called.
For example, take a look at mrjob/examples/mr_spark_wordcount.py. We could run the script like this:
python mr_spark_wordcount.py -r emr -v --image-version 5.10.0 input1.txt input2.txtHere's what would happen in the code:
A code snippet at the bottom of mr_spark_wordcount.py would call MRSparkWordcount.run(), which would itself constructs an instance a MRSparkWordcount (let's call it self).
(All these methods are defined in the base MRJob class; trace through the code in mrjob/job.py.)
The MRJob constructor itself sets up an self.arg_parser and then calls self.configure_args(), which updates self.arg_parser, and self.load_args(), which puts the command-line arguments into self.options.
(Our subclass of MRJob doesn't redefine configure_args() or load_args(), but it's fairly common to do so, especially to add custom command-line switches.)
Next, the run() method calls execute() on the job we just constructed. execute() itself calls self.run_job(), which makes use of the command line arguments to set up the runner.
In this case:
-
-vmeans to use verbose logging (self.options.verbose = True) -
-r emrmeans to run on Elastic MapReduce (self.options.runner = 'emr') -
--image-version 5.10.0means something specific to Elastic MapReduce, in this case, using AMI version 5.10.0 (self.options.image_version = 5.10.0).
self.run_job() first sets up logging:
self.set_up_logging(verbose=self.options.verbose, ...)and then calls self.make_runner(), which uses the command-line options and some knowledge about the job to construct a runner like this:
runner = EMRJobRunner(
image_version=5.10.0,
mr_job_script=/path/to/mrjob/examples/mr_spark_wordcount.py,
stdin=sys.stdin,
steps=[{'jobconf': {}, 'spark_args': [], 'type': 'spark'}],
...
)If you want to dig deeper in the code, EMRJobRunner is chosen by self._runner_class(), and its constructor arguments are chosen by self._runner_kwargs().
In particular, steps comes from self._steps_desc(), which just boils the result of self.steps() down into a simple JSONable format.
Some MRJobs re-define self.steps(), but in this particular case, our job (see mrjob/examples/mr_spark_wordcount.py) just re-defines self.spark(), resulting in a one-step job with a step of type 'spark'.
Finally, run_job() calls runner.run(). In this case, it also prints the results of the job to standard output. We could have repressed this by calling the job with -o s3://bucket/path or --no-cat-output.
Note that MRJobs don't have to run themselves. You can also farm out setting up logging and parsing command-line arguments to an external script, which would then need to do something like this:
job = MRYourJob(args=[...])
with job.make_runner() as runner:
runner.run()
for k, v in job.parse_output(runner.cat_output()):
...If you look at the code, this is actually pretty similar to what self.run_job() does.
We'll get into more of what runner.run() does below, but the gist of it is that it copies mr_spark_wordcount.py (and our input files) into our Spark cluster on EMR, and then calls it with --step-num=0 --spark and the paths of our input files:
spark-submit ... s3://bucket/path/mr_spark_wordcount.py --step-num=0 --spark s3://bucket/path/to/tmp/input1.txt,s3://bucket/path/to/tmp/input2.txt s3://bucket/path/to/tmp/output/Spark will call our script once with the command-line arguments above. The resulting process is called the Spark driver.
The code path for the driver is the same as above (refer back to mrjob/job.py, except now when we get to self.execute() it notices the --spark switch (self.options.spark = True) and instead of calling self.run_job(), it calls self.run_spark(step_num=0). self.run_spark() then calls the spark() method defined in mrjob/examples/mr_spark_wordcount.py with our input and output paths:
self.spark(
's3://bucket/path/input1.txt,s3://bucket/path/input2.txt',
's3://bucket/path/output/'
)At this point, our spark() method creates a SparkContext, and with the help of the pyspark library, creates several copies of itself that run different bits of code on different bits of data.
With Hadoop Streaming jobs, there is no pyspark to help us out, so mrjob is responsible for a bit more of the legwork; there's an example below.
The runner is responsible for getting your job into Spark or Hadoop Streaming and making it run. Runners are always a subclass of MRJobRunner (see mrjob/runner.py).
Unlike with jobs, you don't have to define your runner class; mrjob picks one based on your command line arguments. For example, -r emr corresponds to EMRJobRunner in mrjob/emr.py.
Runners do three things:
- ship files to Spark or Hadoop
- construct command lines to run
- launch and monitor the job
Here's what happens in the code when runner.run() is called:
One of the most helpful things mrjob does is make sure the files you need to run your job are where Spark or Hadoop Streaming needs them. These may include:
- your mrjob script (in this case mr_spark_wordcount.py)
- a copy of the MRJob library
- files specified in your job definition (e.g. with
add_file_arg()or theFILESattribute) - files specified on the command line
- files needed by bootstrap or setup scripts (more about that below)
For example, to run our job in the cloud on EMR, we not only have to instruct EMR to place mr_spark_wordcount.py and a copy of mrjob where Hadoop or Spark can see them, we also have to put a copy of these files somewhere on S3 so EMR can see them.
The MRJobRunner's run() method (in mrjob/runner.py) is where to start when tracing the code. Unfortunately, most of the magic happens in each runner subclass's _run() method (note the underscore), and these very a bit from runner to runner. (See below to learn more about individual runner classes.)
When you run on EMR, the first thing you have to do is make sure that files on the local filesystem that we want the job to access are copied to S3. This is handled by self._upload_mgr, which is an instance of UploadDirManager, defined in mrjob/setup.py. The main job of the UploadDirManager is just to keep track of which files need to be uploaded, and assign them unique names.
Not all runners need to set self._upload_mgr; for example, if you are running a job locally, there is no need to upload files to a remote filesystem. In our case, we're running on EMR, and self._upload_mgr is defined in the constructor of EMRJobRunner, in mrjob/emr.py:
self._upload_mgr = UploadDirManager(s3_files_dir)Next, we need to make sure that files we want to be available to the job appear in the working directory of each task inside Hadoop/Spark. As far as Hadoop and Spark are concerned, there are basically two kinds of things you might want to make available to your job: files (which are basically any type of file, but just one), and archives, which start life as tarballs but get unpacked as directories inside Hadoop and Spark.
Both files and archives are tracked by self._working_dir_mgr, which is defined for all runners, and is an instance of WorkingDirManager (also defined in mrjob/setup.py). WorkingDirManager is fairly similar to UploadDirManager, but it knows how to distinguish between files and archives.
In addition to files and archives, MRJob also supports dirs, which are just archives that MRJob creates for you by tarballing existing directories. These are converted into archives in self._create_dir_archive(), called from self.run() (both of these are in mrjob/runner.py). From the point of view of the WorkingDirManager, dirs are just archives.
Occasionally, you will see other WorkingDirManagers in the code. For example in EMRJobRunner, we also have self._bootstrap_dir_mgr and self._master_node_setup_mgr. For now, just know that these are doing the exact same thing (tracking files and archives to ship into a working directory), just for other sorts of tasks (e.g. bootstrapping the cluster).
Files get added to self._working_dir_mgr and self._upload_mgr all over the code, generally at the point that they are first created or identified. For example, on most runners, we create a copy of the mrjob library (self._create_mrjob_zip(), defined in mrjob/bin.py) and then immediately add it to the relevant runner. This can look a bit like "spaghetti code", but it allows us to separate tracking files (by adding them to the appropriate manager) from uploading them.
The last two things any runner with an upload manager does before actually launching the job are call _add_job_files_for_upload(), which adds file from the runner's WorkingDirManager(s) to self._upload_mgr, and self._upload_local_files() which handles copying files from your local filesystem to the remote filesystem (e.g. S3). add_job_files_for_upload() is defined differently for every runner, but there's only one self._upload_local_files(), defined in mrjob/runner.py).
The actual work of getting files into the job's working directory is handled by Hadoop or Spark; all mrjob does is pass the appropriate command-line arguments to hadoop or spark-submit. Which brings us to...
Let's return to our example, above. We ran this command line:
python mr_spark_wordcount.py -r emr -v --image-version 5.10.0 input1.txt input2.txtOn EMR, it creates a step with this definition:
{
'Jar': 'command-runner.jar',
'Args': [
'spark-submit',
'--conf',
'spark.executorEnv.PYSPARK_PYTHON=python',
'--conf',
'spark.yarn.appMasterEnv.PYSPARK_PYTHON=python',
'--master',
'yarn',
'--deploy-mode',
'cluster',
'--files',
's3://bucket/path/to/tmp/files/wd/mr_spark_wordcount.py',
's3://bucket/path/to/tmp/files/wd/mr_spark_wordcount.py',
'--step-num=0',
'--spark',
's3://bucket/path/to/tmp/files/input1.txt,s3://bucket/path/to/tmp/files/input2.txt',
's3://path/to/tmp/output/'
]
}which instructs EMR to run this command line on the cluster's master node:
spark-submit --conf ... --master yarn --deploy-mode cluster --files ... s3://bucket/path/to/tmp/files/wd/mr_spark_wordcount.py ...```
In this case, mrjob knows that EMR runs Spark jobs in cluster mode on the YARN master (that is, on Hadoop). It's actually a bit overzealous here; it both uploads our Spark script into the working directory, and then tells Spark to run it directly from S3 (which you can do in cluster mode).
This command line is created by `self._args_for_spark_step()`, defined in [mrjob/bin.py](https://github.com/Yelp/mrjob/blob/master/mrjob/bin.py)). This in turn calls various methods defined in the same module:
```python
def _args_for_spark_step(self, step_num, last_step_num=None):
return (
self.get_spark_submit_bin() +
self._spark_submit_args(step_num) +
[self._spark_script_path(step_num)] +
self._spark_script_args(step_num, last_step_num)
)Some other switches that _spark_submit_args() could potentially add to the command line include:
- More
--confswitches, either from:- spark properties set with
--jobconf(e.g.spark.executor.memory) - environment variables set with
--cmdenv(like we do above forPYSPARK_PYTHON)
- spark properties set with
- An
--archivesswitch for archives that are meant to be uploaded to the working directory -
--py-files, for Python packages added (set with mrjob's own--py-filesswitch)
It's pretty common for mrjob to run Spark scripts with --py-files /path/to/mrjob.zip, ensuring they have access to a copy of the mrjob library. The only reason this doesn't happen on EMR is that we install a copy of mrjob on the cluster at bootstrap time (see below).
Finally, we need to actually run the job and wait until it finishes. On EMR this consists of four steps:
- a. create a cluster
- b. submit steps
- c. wait for the steps to finish
- d. parse logs if the job failed
For most runners, steps a and b are replaced by simply running a command (spark-submit ... or hadoop jar ...)
Running the command-line above will cause mrjob to make an EMR API call like this:
RunJobFlow(
Applications=[{'Name': 'Hadoop'}, {'Name': 'Spark'}],
BootstrapActions=[
{'Name': 'master', 'ScriptBootstrapAction': {'Path': 's3://bucket/path/to/tmp/files/b.sh', 'Args': []}},
],
Instances={'InstanceGroups': [...],
JobFlowRole='mrjob-...',
LogUri='s3://bucket/path/to/logs/',
Name='mr_spark_wordcount.user.timestamp',
ReleaseLabel='emr-5.10.0',
ServiceRole='mrjob-...',
Steps=[]
)Some things to note:
-
Applications: mrjob automatically determines we need to installSpark -
Instances: mrjob configures how many and what type of EC2 instances, based on user request -
LogUri: mrjob always requests logs be copied to S3 -
JobFlowRoleandServiceRole: if you don't configure these, mrjob takes care of setting IAM up for you -
ReleaseLabel: set based on our--image-version 5.10.0on the command line -
Steps: empty because we will do this later
Finally, BootstrapActions tell EMR to run a script on each host as root before running any steps. In this case, it's a script that installs mrjob, and it looks like this:
#!/bin/sh -x
set -e
# store $PWD
__mrjob_PWD=$PWD
if [ $__mrjob_PWD = "/" ]; then
__mrjob_PWD=""
fi
{
# download files and mark them executable
aws s3 cp s3://mrjob-35cdec11663cb1cb/tmp/mr_spark_wordcount.marin.20191204.222645.633132/files/mrjob.zip $__mrjob_PWD/mrjob.zip
chmod u+rx $__mrjob_PWD/mrjob.zip
# bootstrap commands
__mrjob_PYTHON_LIB=$(python3 -c 'from distutils.sysconfig import get_python_lib; print(get_python_lib())')
sudo rm -rf $__mrjob_PYTHON_LIB/mrjob
sudo unzip $__mrjob_PWD/mrjob.zip -d $__mrjob_PYTHON_LIB
sudo python3 -m compileall -q -f $__mrjob_PYTHON_LIB/mrjob && true
} 1>&2As noted above, bootstrap scripts have their own instance of WorkingDirMgr. Its contents become commands to download the file (aws s3 cp ...), and, for archives, to un-archive them as well (tar ...).
mrjob would then make an EMR API call like this:
AddJobFlowSteps(
JobFlowId='j-CLUSTERID',
Steps=[
{
'ActionOnFailure': 'TERMINATE_CLUSTER',
'HadoopJarStep': {
'Jar': 'command-runner.jar',
'Args': [...]
},
'Name': 'mr_spark_wordcount.user.timestamp: Step 1 of 1'
}
]
)(We covered the contents of Args above.)
This step definition is created by self._build_step() in mrjob/emr.py.
mrjob makes repeated DescribeCluster and DescribeStep API calls every 30 seconds or so, until your step completes or fails, or the cluster gets shut down. You can trace through the code starting at self._wait_for_steps_to_complete() in in mrjob/emr.py
If the job completes successfully, run() is done. At that point, your MRJob class (or your own batch script) might call runner.cat_output().
Once your runner goes out of scope, runner.cleanup() will be called, causing temp directories to be deleted and so on. EMR will automatically shut down the cluster once it runs out of steps. (There is a way for mrjob to launch a cluster that EMR won't automatically shut down, but in that case, mrjob adds a special bootstrap script that shuts the cluster down after it's been idle for a certain number of minutes.)
One of the cool things about mrjob is that it's pretty good at finding the error when your job fails.
How the log parsing code works is a whole other topic, but essentially mrjob will very selectively download and parse logs from S3 (or, if possible, by SSHing into your cluster) until it finds the Python exception that probably caused the job to fail. (If it can't find one, it eventually gives up and prints a Java error instead.)
The entry point into log parsing is self._pick_error(), defined in LogInterpretationMixin in mrjob/logs/mixin.py.
Once mrjob is done parsing the logs, it'll throw a StepFailedException.
The examples above use a Spark job, but mrjob was actually originally was designed to work with Hadoop. Here are some other kinds of jobs that mrjob can run, and what they look like in terms of code and command lines.
Back in 2010 when mrjob was first released, there was no mainstream big data solution that used Python (Spark wasn't released until 2014). Instead, mrjob made use of Hadoop Streaming, a language-neutral solution.
Hadoop Streaming jobs generally operate on lines of data, which may be spread across multiple input files. A Hadoop Streaming job has no more than one mapper command, and no more than one reducer command. These commands take in lines through standard input, and output lines through standard output.
First data is divided up and the mapper command is run on each. Then the resulting lines are shuffled and sorted by key (which is just the part of the line before the first TAB character). Finally, the shuffled and sorted lines are divided up between reducers in such a way that all lines with the same key are adjacent and are processed by the same reducer.
Just as a Spark MRJob can run inside Spark, a MRJob designed for Hadoop streaming can serve as both mapper and reducer inside Hadoop.
As an example, let's MRWordFreqCount, a simple word frequency count job, in mrjob/examples/mr_word_freq_count.py:
class MRWordFreqCount(MRJob):
def mapper(self, _, line):
for word in WORD_RE.findall(line):
yield (word.lower(), 1)
def reducer(self, word, counts):
yield (word, sum(counts))The mapper() method is concerned with finding the words in a line. For example, a mapper fed "Go Dog, Go!" as an input line would emit: ("go", 1), ("dog", 1), ("go", 1).
The job of the reducer() method is to collect all the words and sum together their counts. For example, reducer() here will be called exactly once with word set to "go" and counts being a stream of 1s (at least two, plus some from other lines).
If you're reading the source code of the job, you may notice there is also a combiner() method. A combiner is essentially an optional mini-reducer that runs before the shuffle-and-sort step. Essentially, several mappers will process data on one node (one computer, basically), and the combiner can reduce the size of the data on that one node before shipping it off to others.
If you run mr_word_freq_count.py on EMR (or some other Hadoop cluster):
python mr_word_freq_count.py -r emr -v input1.txt input2.txtmrjob will construct a command line something like this:
hadoop jar /path/to/streaming.jar -mapper 'python mr_word_freq_count.py --step-num=0 --mapper' -combiner 'python mr_word_freq_count.py --step-num=0 --combiner' -reducer 'python mr_word_freq_count.py --step-num=0 --reducer' -files s3://bucket/path/to/tmp/mr_word_freq_count.py -input s3://bucket/path/to/tmp/input1.txt -input bucket/path/to/tmp/input2.txt -output s3://bucket/path/to/tmp/output/ Essentially, rather than our script being run with --spark, it's run as a mapper (--mapper), combiner (--combiner) and reducer (-reducer). And because of the way Hadoop Streaming works, our script literally gets run from start to finish many, many times across different nodes.
This command line to run Hadoop is created by self._hadoop_streaming_jar_args() in mrjob/bin.py. Most of the methods it calls are in that file as well.
When Hadoop Streaming runs your script with --mapper, run_mapper() in mrjob/job.py gets called, and it in turn runs your mapper method once for each line.
Similarly, when Hadoop Streaming runs your script with --reducer, run_reducer() gets called, and your reducer() method gets run once for each key. If you trace through the code, you'll see that mrjob ultimately uses itertools.groupby() to present all the values corresponding to one key as a stream of data.
From your MRJob's point of view, combiners work basically the same way as reducers (script gets called with --combiner, calls run_combiner()).
Unlike Spark, which is aware of Python data structures, Hadoop Streaming just operates on lines. So mrjob offers protocols to convert Python data structures to and from lines.
By default, the initial mapper receives the raw line, and from then on out, everything is encoded as JSON. You can change this by setting INPUT_PROTOCOL, OUTPUT_PROTOCOL, and/or INTERNAL_PROTOCOL to a protocol class in your MRJob class definition. You can see several protocols in mrjob/protocol.py; for example, you can use pickle instead of JSON.
Hadoop Streaming can run any command as a mapper or reducer, not just your MRJob script. You can leverage this by re-defining mapper_cmd() or reducer_cmd() (or for that matter, combiner_cmd()). For example, mrjob/examples/mr_grep.py does this. When you run the script, it'll make a command line like:
hadoop jar streaming.jar -mapper 'grep -e <expression>'Running a single mapper and reducer isn't all that useful for most tasks, so mrjob allows you to create multi-step jobs. From the point of view of Hadoop or Spark, these are just a series of jobs where the output directory of one step happens to be the same as the input directory of the next step.
To define a multi-step MRJob, you re-define self.steps(). For example, here's the steps() method for MRMostUsedWord (in mrjob/examples/mr_most_used_word.py):
def steps(self):
return [
MRStep(mapper_init=self.mapper_init,
mapper=self.mapper_get_words,
combiner=self.combiner_count_words,
reducer=self.reducer_count_words),
MRStep(reducer=self.reducer_find_max_word)
]MRStep is just the class that represents a Hadoop Streaming step; you can see other step classes (including SparkStep) in mrjob/step.py.
The command lines for multi-step jobs are constructed in exactly the same way, except that commands from the second step are run with --step-num=1 (so running mr_most_used_word.py
You might notice mapper_init=.... This is just a method that takes no arguments that runs once every time the mapper script is called; in this script we use it to load a file containing stop words.
It's possible to combine two different kinds of steps in a single job; for example, see mrjob/examples/mr_jar_step_example.py
By the way, if you don't redefine self.steps(), all that happens is that MRJob's default step method (see mrjob/job.py) checks which other methods have been re-defined and returns an array. For example, if you re-define mapper(), the default steps() method returns [MRStep(mapper=self.mapper, ...)].
Hadoop Streaming is actually just one of many JARs you can run. If you have good knowledge of Hadoop and Java, you can actually write your own JARs, which potentially run much faster.
If you define a MRJob with a JarStep, it just constructs a command line like hadoop jar your.jar <args>. It's also up to you to decide what, if any arguments your JAR receives. You can use the constants INPUT, OUTPUT, and GENERIC_ARGS to stand for the input files or directory, the output directory, and any generic Hadoop arguments (e.g. -D for configuration properties). Here's some example code from mrjob/examples/mr_jar_step_example.py that constructs a JarStep:
jar_step = JarStep(
jar='file:///usr/lib/hadoop-mapreduce/hadoop-mapreduce-examples.jar',
args=['wordcount', GENERIC_ARGS, INPUT, OUTPUT],
)When mrjob runs a JarStep, it doesn't need to upload your MRJob script. If the JAR is a local file (not a URI as above), it will ensure that the JAR gets uploaded.
Similarly, spark-submit can run any Python script or Spark JAR, not just MRJob scripts. You can run a regular old Spark script by defining a SparkScriptStep, and similarly, you can run a Spark JAR by defining a SparkJarStep.
If all you want to do is run a Spark script on EMR or some other platform mrjob supports, you don't even need to make a script; you can just run mrjob spark-submit ... as a drop-in replacement for spark-submit (code for this is in mrjob/tools/spark_submit.py).
Hadoop Streaming jobs can set Hadoop input and output formats, in order to read input or write output in special ways.
For example, mrjob/examples/mr_nick_nack.py is basically a word frequency count job, but it sets HADOOP_OUTPUT_FORMAT = 'nicknack.MultipleValueOutputFormat' so that output is divided into separate directories based on the first letter of the word. It also sets LIBJARS to ensure that the JAR containing MultipleValueOutputFormat is automatically uploaded into Hadoop Streaming.
Sometimes you want to run a job that processes files rather than lines. MRJob makes this easy for you; just define mapper_raw() in your job (or make a step like MRStep(mapper_raw=...)) and it'll pass the path of a local copy of the file, and its URI on S3 or wherever it's stored.
For example, mrjob/examples/mr_phone_to_url.py uses this feature to parse WARC files, which encode webpages using a multi-line format.
What's going on behind the scenes is that the "input" that Hadoop Streaming sees is a list of URIs of files, and then it sets your script's Hadoop input format to NLineInputFormat, so that each mapper only receives the URI of one file.
Your script still gets called with --mapper --step-num=...; just run_mapper() does something slightly different when the script runs.
You can add your own custom switches to your job, and they'll get passed through to your script when it runs inside Spark or Hadoop.
For example, mrjob/examples/mr_text_classifier.py uses self.add_passthru_arg() to add a --min-df (document frequency) switch to the job:
def configure_args(self):
super(MRTextClassifier, self).configure_args()
self.add_passthru_arg(
'--min-df', dest='min_df', default=2, type=int,
help=('min number of documents an n-gram must appear in for us to'
' count it. Default: %(default)s'))
...Now, the commands that Hadoop Streaming runs will include --min-df (-mapper 'mr_text_classifier.py --mapper --step-num=0 --min-df=1) if you set it. This works with SparkSteps as well—one reason not to just write a regular Spark script.
You can also add special switches that take files as a single argument; if the file is on your local filesystem, mrjob will ensure that it gets uploaded into Spark or Hadoop. For example, mrjob/examples/mr_most_used_word.py uses this to upload a custom stop words file:
def configure_args(self):
super(MRMostUsedWord, self).configure_args()
self.add_file_arg(
'--stop-words-file',
dest='stop_words_file',
default=None,
help='alternate stop words file. lowercase words, one per line',
)So far we've only talked about running jobs on EMR. In addition to the EMR runner (in mrjob/emr.py), there are five others:
InlineMRJobRunner (in mrjob/inline.py) avoids launching subprocesses from Python, to make debugging easier. If you run a Hadoop Streaming job, it does its best to simulate a single-threaded version of Hadoop (it can't run Hadoop JARs though). If you run a Spark job, it actually runs your job through the pyspark library.
If you don't specify the -r flag, this is the runner you get by default, on the assumption that you'd probably rather debug your job before running it on a cluster.
LocalMRJobRunner (in mrjob/local.py) is like InlineMRJobRunner, except that it launches subprocesses. When running Hadoop Streaming jobs, it's essentially a poor man's Hadoop; it uses all your CPUs, but has no error recovery. In addition, when launching a Spark job, it uses a local-cluster[...] Spark master, which is a better way to test out complex jobs locally.
The Hadoop runner (in mrjob/hadoop.py) is designed to work with your own Hadoop cluster. If the hadoop binary is in one of about six standard places, it'll find it and start launching hadoop jar and hadoop fs commands (for HDFS). It runs Spark as well, assuming by default you'll want to use the yarn master and HDFS.
SparkMRJobRunner (in mrjob/spark/runner.py) always launches jobs spark-submit. What's cool about this runner is that it can run Hadoop Streaming jobs as well, but it runs them in Spark! (More below about the magic behind this.)
The Spark runner is, like Spark, agnostic about filesystems, and supports any filesystem supported by mrjob (local, HDFS, S3, or Google Cloud Storage).
DataprocJobRunner (in mrjob/dataproc.py) is like EMRJobRunner, except that instead of working with Amazon's Elastic MapReduce, it runs jobs on Google Cloud Dataproc.
Unfortunately, DataprocJobRunner does not yet support Spark.
Here's what the entire runner class hierarchy looks like:
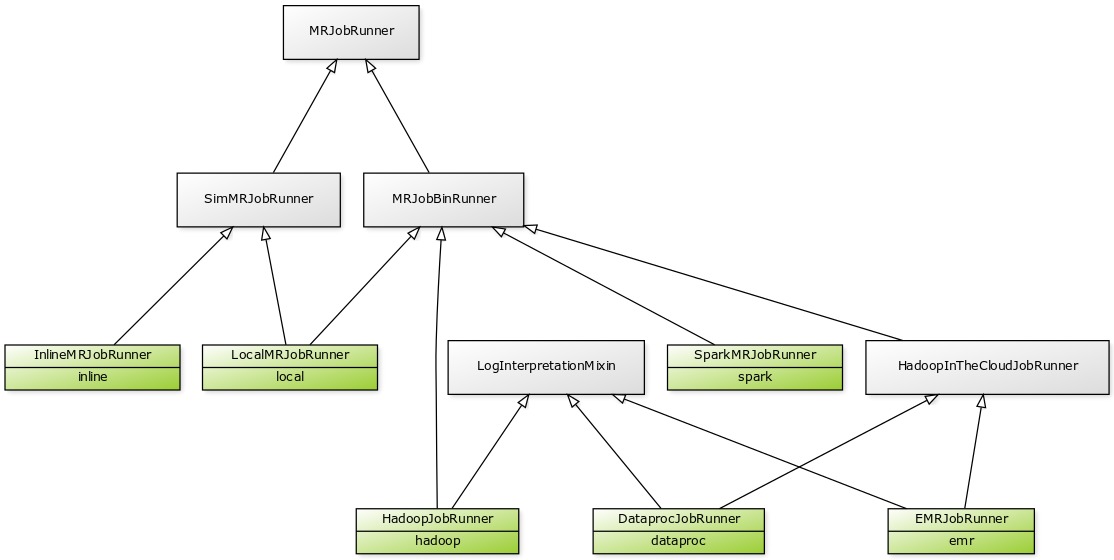
If you're trying to make a change to runner code, here's a rough guide to where to look:
BinMRJobRunner (in mrjob/bin.py) contains the bulk of shared runner code that involves launching subprocesses or creating command lines. Basically, if every runner but InlineMRJobRunner needs a certain feature, you'll find it here.
Code common to all runners is found in the base MRJobRunner class (in mrjob/runner.py).
SimMRJobRunner (in mrjob/sim.py) contains nearly all the code that powers -r inline and -r local. This handles running mappers and reducers, setting up fake Hadoop working directories and configuration properties, etc.
LogInterpretationMixin (in mrjob/logs/mixin.py) handles downloading and parsing logs created by the job. Note that SparkMRJobRunner doesn't inherit from LogInterpretationMixin because it only parses the output of the spark-submit command, which it parse directly with other code in mrjob/logs/.
HadoopInTheCloudJobRunner (in mrjob/cloud.py) contains some code that's specifically useful for cloud services (launching clusters, SSHing in, etc.). There is doubtless still some common code between EMRJobRunner and DataprocJobRunner that could be pushed up into this class but hasn't yet been.
As noted above, the mrjob library is older than Spark. What are users supposed to do with their old Hadoop Streaming MRJob scripts if they migrate to a Hadoop-free Spark solution, such as Spark on Mesos?
Hadoop Streaming MRJob scripts are essentially just a description, in Python, of how to transform data. It turns out that everything you can do in Hadoop Streaming, you can also do in Spark.
When MRJobSparkRunner wants to run a Hadoop Streaming job, the Spark script it actually runs is mrjob/spark/harness.py, which we call the Spark harness. This script just needs to know the name of your MRJob class, and simplified JSON version of your job's steps, which MRJobSparkRunner passes through as part of harness.py's command line.
At this point the harness calls your job's mappers, reducers, etc. It doesn't actually call run_mapper(), but instead calls map_pairs() a method that run_mapper() calls (see mrjob/job.py). Similarly, there are reduce_pairs() and combine_pairs().
As magical as it is, the source code for mrjob/spark/harness.py is pretty straightforward; it's only long because it covers nearly every edge case needed by real MRJobs.
The main thing that's tricky is that Hadoop's concept of a reducer is much more heavyweight than Spark's, so we actually use rdd.mapPartitions() to simulate reducers. Hadoop's combiners are a better fit for Spark's lightweight paradigm, so there's a special code path (using rdd.combineByKey()) to make use of combiners if they're defined.
In general, Hadoop Streaming MRJobs tend to run faster in the Spark harness than in actual Hadoop Streaming. Hadoop Streaming retains two main advantages:
- it can run arbitrary commands (e.g.
grep). (Spark probably could do this, but then we'd be literally turning Spark into Hadoop Streaming.) - it has no limit on reducer size. In Hadoop Streaming, all the values for one reducer key go into a file, and as long as your job doesn't load them all into memory, it can handle arbitrarily many. In contrast, Spark is all about loading stuff into memory, so if you have a particularly "lumpy" dataset, Spark may choke on it before your code gets to run.